Note
Click here to download the full example code or to run this example in your browser via Binder
Dirty categories: machine learning with non normalized strings¶
Including strings that represent categories often calls for much data preparation. In particular categories may appear with many morphological variants, when they have been manually input or assembled from diverse sources.
Here we look at a dataset on wages [1] where the column ‘Employee Position Title’ contains dirty categories. On such a column, standard categorical encodings leads to very high dimensions and can lose information on which categories are similar.
We investigate various encodings of this dirty column for the machine learning workflow, predicting the ‘Current Annual Salary’ with gradient boosted trees. First we manually assemble a complex encoder for the full dataframe, after which we show a much simpler way, albeit with less fine control.
The data¶
We first retrieve the dataset:
from dirty_cat.datasets import fetch_employee_salaries
employee_salaries = fetch_employee_salaries()
X, the input data (descriptions of employees):
X = employee_salaries.X
X
and y, our target column (the annual salary):
y = employee_salaries.y
y.name
Out:
'current_annual_salary'
Now, let’s carry out some basic preprocessing:
import pandas as pd
X["date_first_hired"] = pd.to_datetime(X["date_first_hired"])
X["year_first_hired"] = X["date_first_hired"].apply(lambda x: x.year)
# Get a mask of the rows with missing values in 'gender'
mask = X.isna()["gender"]
# And remove them
X.dropna(subset=["gender"], inplace=True)
y = y[~mask]
Assembling a machine-learning pipeline that encodes the data¶
To build a learning pipeline, we need to assemble encoders for each column, and apply a supervised learning model on top.
The categorical encoders¶
An encoder is needed to turn a categorical column into a numerical representation:
from sklearn.preprocessing import OneHotEncoder
one_hot = OneHotEncoder(handle_unknown="ignore", sparse=False)
We assemble these to apply them to the relevant columns.
The ColumnTransformer is created by specifying a set of transformers
alongside with the column names on which each must be applied:
from sklearn.compose import make_column_transformer
encoder = make_column_transformer(
(one_hot, ["gender", "department_name", "assignment_category"]),
("passthrough", ["year_first_hired"]),
# Last but not least, our dirty column
(one_hot, ["employee_position_title"]),
remainder="drop",
)
Pipelining an encoder with a learner¶
We will use a HistGradientBoostingRegressor,
which is a good predictor for data with heterogeneous columns
(we need to require the experimental feature for scikit-learn versions
earlier than 1.0):
from sklearn.experimental import enable_hist_gradient_boosting
# We can now import the |HGBR| from ensemble
from sklearn.ensemble import HistGradientBoostingRegressor
# We then create a pipeline chaining our encoders to a learner
from sklearn.pipeline import make_pipeline
pipeline = make_pipeline(encoder, HistGradientBoostingRegressor())
Out:
/usr/lib/python3/dist-packages/sklearn/experimental/enable_hist_gradient_boosting.py:16: UserWarning: Since version 1.0, it is not needed to import enable_hist_gradient_boosting anymore. HistGradientBoostingClassifier and HistGradientBoostingRegressor are now stable and can be normally imported from sklearn.ensemble.
warnings.warn(
The pipeline can be readily applied to the dataframe for prediction:
pipeline.fit(X, y)
Out:
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
Dirty-category encoding¶
The OneHotEncoder is actually not well suited to the ‘Employee
Position Title’ column, as this column contains 400 different entries:
import numpy as np
np.unique(y)
Out:
array([ 9196. , 11147.24, 13244.5 , ..., 233003. , 239566. ,
303091. ])
We will now experiment with encoders specially made for handling dirty columns:
from dirty_cat import (
SimilarityEncoder,
TargetEncoder,
MinHashEncoder,
GapEncoder,
)
encoders = {
"one-hot": one_hot,
"similarity": SimilarityEncoder(),
"target": TargetEncoder(handle_unknown="ignore"),
"minhash": MinHashEncoder(n_components=100),
"gap": GapEncoder(n_components=100),
}
We now loop over the different encoding methods,
instantiate a new Pipeline each time, fit it
and store the returned cross-validation score:
from sklearn.model_selection import cross_val_score
all_scores = dict()
for name, method in encoders.items():
encoder = make_column_transformer(
(one_hot, ["gender", "department_name", "assignment_category"]),
("passthrough", ["year_first_hired"]),
# Last but not least, our dirty column
(method, ["employee_position_title"]),
remainder="drop",
)
pipeline = make_pipeline(encoder, HistGradientBoostingRegressor())
scores = cross_val_score(pipeline, X, y)
print(f"{name} encoding")
print(f"r2 score: mean: {np.mean(scores):.3f}; std: {np.std(scores):.3f}\n")
all_scores[name] = scores
Out:
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
one-hot encoding
r2 score: mean: 0.776; std: 0.028
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
similarity encoding
r2 score: mean: 0.923; std: 0.014
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
target encoding
r2 score: mean: 0.842; std: 0.030
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
minhash encoding
r2 score: mean: 0.919; std: 0.012
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/lib/python3/dist-packages/sklearn/preprocessing/_encoders.py:828: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
gap encoding
r2 score: mean: 0.921; std: 0.018
Plotting the results¶
Finally, we plot the scores on a boxplot:
import seaborn
import matplotlib.pyplot as plt
plt.figure(figsize=(4, 3))
ax = seaborn.boxplot(data=pd.DataFrame(all_scores), orient="h")
plt.ylabel("Encoding", size=20)
plt.xlabel("Prediction accuracy ", size=20)
plt.yticks(size=20)
plt.tight_layout()
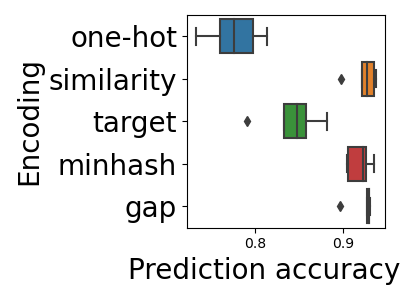
The clear trend is that encoders grasping similarities between categories
(SimilarityEncoder, MinHashEncoder, and GapEncoder) perform better than those discarding it.
SimilarityEncoder is the best performer, but it is less scalable on big
data than the MinHashEncoder and GapEncoder. The most scalable encoder is
the MinHashEncoder. On the other hand, the GapEncoder has the benefit of
providing interpretable features
(see Investigating and interpreting dirty categories)
A simpler way: automatic vectorization¶
The code to assemble a column transformer is a bit tedious. We will now explore a simpler, automated, way of encoding the data.
Let’s start again from the raw data:
employee_salaries = fetch_employee_salaries()
X = employee_salaries.X
y = employee_salaries.y
We’ll drop the ‘date_first_hired’ column as it’s redundant with ‘year_first_hired’.
X = X.drop(["date_first_hired"], axis=1)
We still have a complex and heterogeneous dataframe:
X
The TableVectorizer can to turn this dataframe into a form suited for
machine learning.
Using the TableVectorizer in a supervised-learning pipeline¶
Assembling the TableVectorizer in a Pipeline with a powerful learner,
such as gradient boosted trees, gives a machine-learning method that
can be readily applied to the dataframe.
The TableVectorizer requires at least dirty_cat 0.2.0.
from dirty_cat import TableVectorizer
pipeline = make_pipeline(
TableVectorizer(auto_cast=True), HistGradientBoostingRegressor()
)
Let’s perform a cross-validation to see how well this model predicts:
from sklearn.model_selection import cross_val_score
scores = cross_val_score(pipeline, X, y, scoring="r2")
print(f"scores={scores}")
print(f"mean={np.mean(scores)}")
print(f"std={np.std(scores)}")
Out:
scores=[0.91700051 0.90032882 0.93300376 0.93355301 0.93739009]
mean=0.9242552370239272
std=0.013860836102927439
The prediction performed here is pretty much as good as above but the code here is much simpler as it does not involve specifying columns manually.
Analyzing the features created¶
Let us perform the same workflow, but without the Pipeline, so we can
analyze the TableVectorizer’s mechanisms along the way.
table_vec = TableVectorizer(auto_cast=True)
We split the data between train and test, and transform them:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.15, random_state=42
)
X_train_enc = table_vec.fit_transform(X_train, y_train)
X_test_enc = table_vec.transform(X_test)
The encoded data, X_train_enc and X_test_enc are numerical arrays:
X_train_enc
Out:
array([[0.00000000e+00, 1.00000000e+00, 0.00000000e+00, ...,
7.65077591e-02, 7.76393234e-02, 2.00700000e+03],
[1.00000000e+00, 0.00000000e+00, 0.00000000e+00, ...,
5.00000000e-02, 5.00000000e-02, 2.00500000e+03],
[1.00000000e+00, 0.00000000e+00, 0.00000000e+00, ...,
5.00000000e-02, 5.00000000e-02, 2.00900000e+03],
...,
[1.00000000e+00, 0.00000000e+00, 0.00000000e+00, ...,
5.00000000e-02, 5.00000000e-02, 1.99000000e+03],
[0.00000000e+00, 1.00000000e+00, 0.00000000e+00, ...,
5.00000000e-02, 5.00000000e-02, 2.01200000e+03],
[1.00000000e+00, 0.00000000e+00, 0.00000000e+00, ...,
5.00000000e-02, 5.00000000e-02, 2.01400000e+03]])
They have more columns than the original dataframe, but not much more:
X_train.shape, X_train_enc.shape
Out:
((7843, 8), (7843, 169))
Inspecting the features created¶
The TableVectorizer assigns a transformer for each column. We can inspect this
choice:
from pprint import pprint
pprint(table_vec.transformers_)
Out:
[('low_card_cat',
OneHotEncoder(drop='if_binary', handle_unknown='ignore'),
['gender', 'department', 'department_name', 'assignment_category']),
('high_card_cat',
GapEncoder(n_components=30),
['division', 'employee_position_title', 'underfilled_job_title']),
('remainder', 'passthrough', ['year_first_hired'])]
This is what is being passed to the ColumnTransformer under the hood.
If you’re familiar with how the latter works, it should be very intuitive.
We can notice it classified the columns ‘gender’ and ‘assignment_category’
as low cardinality string variables.
A OneHotEncoder will be applied to these columns.
The vectorizer actually makes the difference between string variables
(data type object and string) and categorical variables
(data type category).
Next, we can have a look at the encoded feature names.
Before encoding:
X.columns.to_list()
Out:
['gender', 'department', 'department_name', 'division', 'assignment_category', 'employee_position_title', 'underfilled_job_title', 'year_first_hired']
After encoding (we only plot the first 8 feature names):
feature_names = table_vec.get_feature_names_out()
feature_names[:8]
Out:
['gender_F', 'gender_M', 'gender_nan', 'department_BOA', 'department_BOE', 'department_CAT', 'department_CCL', 'department_CEC']
As we can see, it gave us interpretable columns.
This is because we used the GapEncoder on the column ‘division’,
which was classified as a high cardinality string variable.
(default values, see TableVectorizer’s docstring).
In total, we have a reasonable number of encoded columns:
len(feature_names)
Out:
169
Feature importances in the statistical model¶
In this section, we will train a regressor, and plot the feature importances.
Note:
To minimize computation time, we use the feature importances computed by the
RandomForestRegressor, but you should prefer permutation_importance()
instead (which are less subject to biases).
First, let’s train the RandomForestRegressor:
from sklearn.ensemble import RandomForestRegressor
regressor = RandomForestRegressor()
regressor.fit(X_train_enc, y_train)
Retrieving the feature importances:
importances = regressor.feature_importances_
std = np.std([tree.feature_importances_ for tree in regressor.estimators_], axis=0)
indices = np.argsort(importances)
# Sort from least to most
indices = list(reversed(indices))
Plotting the results:
import matplotlib.pyplot as plt
plt.figure(figsize=(12, 9))
plt.title("Feature importances")
n = 20
n_indices = indices[:n]
labels = np.array(feature_names)[n_indices]
plt.barh(range(n), importances[n_indices], color="b", yerr=std[n_indices])
plt.yticks(range(n), labels, size=15)
plt.tight_layout(pad=1)
plt.show()
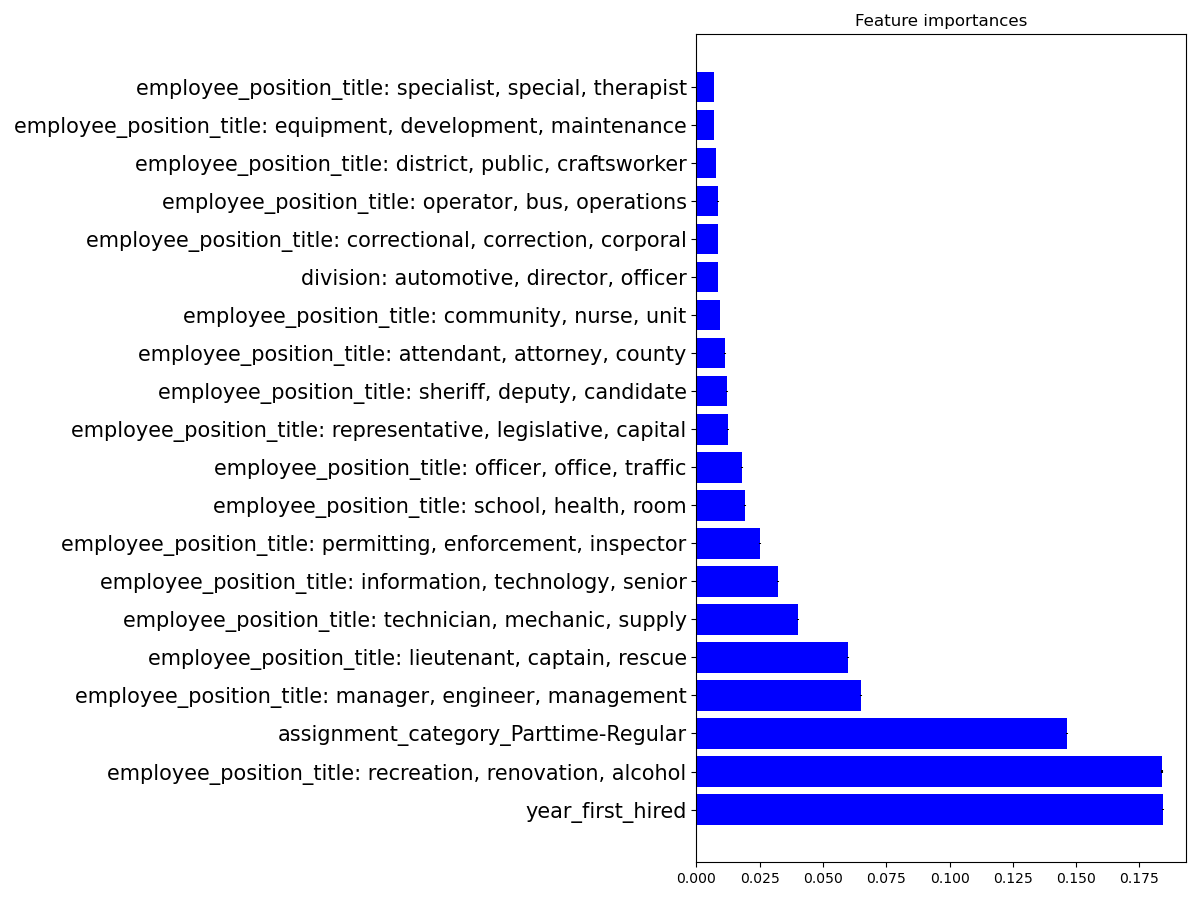
We can deduce from this data that the three factors that define the most the salary are: being hired for a long time, being a manager, and having a permanent, full-time job :)
The TableVectorizer automates preprocessing
As this notebook demonstrates, many preprocessing steps can be
automated by the TableVectorizer, and the resulting pipeline can still be
inspected, even with non-normalized entries.
Total running time of the script: ( 3 minutes 35.437 seconds)