Note
Click here to download the full example code or to run this example in your browser via Binder
Fuzzy joining dirty tables and the FeatureAugmenter¶
Here we show how to combine data from different sources, with a vocabulary not well normalized.
Joining is difficult: one entry on one side does not have an exact match on the other side.
The fuzzy_join() function enables to join tables without cleaning the data by
accounting for the label variations.
To illustrate, we will join data from the 2022 World Happiness Report. with tables provided in the World Bank open data platform in order to create a first prediction model.
Moreover, the FeatureAugmenter() is a scikit-learn Transformer that makes it easy to
use such fuzzy joining multiple tables to bring in information in a
machine-learning pipeline. In particular, it enables tuning parameters of
fuzzy_join() to find the matches that maximize prediction accuracy.
Data Importing and preprocessing¶
We import the happiness score table first:
import pandas as pd
df = pd.read_csv(
"https://raw.githubusercontent.com/dirty-cat/datasets/master/data/Happiness_report_2022.csv",
thousands=",",
)
df.drop(df.tail(1).index, inplace=True)
Let’s look at the table:
df.head(3)
This is a table that contains the happiness index of a country along with some of the possible explanatory factors: GDP per capita, Social support, Generosity etc.
For the sake of this example, we only keep the country names and our variable of interest: the ‘Happiness score’.
df = df[["Country", "Happiness score"]]
Additional tables from other sources¶
Now, we need to include explanatory factors from other sources, to complete our covariates (X table).
Interesting tables can be found on the World Bank open data platform, for which we have a downloading function:
from dirty_cat.datasets import fetch_world_bank_indicator
We extract the table containing GDP per capita by country:
gdppc = fetch_world_bank_indicator(indicator_id="NY.GDP.PCAP.CD").X
gdppc.head(3)
Then another table, with life expectancy by country:
life_exp = fetch_world_bank_indicator("SP.DYN.LE00.IN", "life_exp").X
life_exp.head(3)
And a table with legal rights strength by country:
legal_rights = fetch_world_bank_indicator("IC.LGL.CRED.XQ").X
legal_rights.head(3)
A correspondance problem¶
Alas, the entries for countries do not perfectly match between our original table (df), and those that we downloaded from the worldbank (gdppc):
df.sort_values(by="Country").tail(7)
gdppc.sort_values(by="Country Name").tail(7)
We can see that Yemen is written “Yemen*” on one side, and “Yemen, Rep.” on the other.
We also have entries that probably do not have correspondances: “World” on one side, whereas the other table only has country-level data.
Joining tables with imperfect correspondance¶
We will now join our initial table, df, with the 3 additional ones that we have extracted.
1. Joining GDP per capita table¶
To join them with dirty_cat, we only need to do the following:
from dirty_cat import fuzzy_join
# We will ignore the warnings:
import warnings
warnings.filterwarnings("ignore")
df1 = fuzzy_join(
df, # our table to join
gdppc, # the table to join with
left_on="Country", # the first join key column
right_on="Country Name", # the second join key column
return_score=True,
)
df1.tail(20)
# We merged the first WB table to our initial one.
We see that our fuzzy_join() succesfully identified the countries,
even though some country names differ between tables.
For instance, “Czechia” is well identified as “Czech Republic” and “Luxembourg*” as “Luxembourg”.
Let’s do some more inspection of the merging done.
Let’s print the four worst matches, which will give us an overview of the situation:
df1.sort_values("matching_score").head(4)
We see that some matches were unsuccesful (e.g “Palestinian Territories*” and “Palau”), because there is simply no match in the two tables.
In this case, it is better to use the threshold parameter so as to include only precise-enough matches:
df1 = fuzzy_join(
df,
gdppc,
left_on="Country",
right_on="Country Name",
match_score=0.35,
return_score=True,
)
df1.sort_values("matching_score").head(4)
Matches that are not available (or precise enough) are marked as NaN. We will remove them using the drop_unmatched parameter:
df1 = fuzzy_join(
df,
gdppc,
left_on="Country",
right_on="Country Name",
match_score=0.35,
drop_unmatched=True,
)
df1.drop(columns=["Country Name"], inplace=True)
We can finally plot and look at the link between GDP per capital and happiness:
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_context("notebook")
plt.figure(figsize=(4, 3))
ax = sns.regplot(
data=df1,
x="GDP per capita (current US$)",
y="Happiness score",
lowess=True,
)
ax.set_ylabel("Happiness index")
ax.set_title("Is a higher GDP per capita linked to happiness?")
plt.tight_layout()
plt.show()
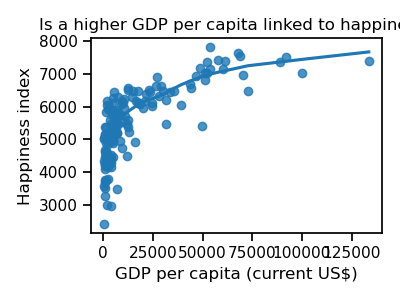
It seems that the happiest countries are those having a high GDP per capita. However, unhappy countries do not have only low levels of GDP per capita. We have to search for other patterns.
2. Joining life expectancy table¶
Now let’s include other information that may be relevant, such as in the life_exp table:
df2 = fuzzy_join(
df1,
life_exp,
left_on="Country",
right_on="Country Name",
match_score=0.45,
)
df2.drop(columns=["Country Name"], inplace=True)
df2.head(3)
Let’s plot this relation:
plt.figure(figsize=(4, 3))
fig = sns.regplot(
data=df2,
x="Life expectancy at birth, total (years)",
y="Happiness score",
lowess=True,
)
fig.set_ylabel("Happiness index")
fig.set_title("Is a higher life expectancy linked to happiness?")
plt.tight_layout()
plt.show()
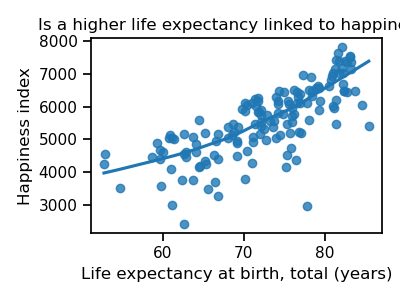
It seems the answer is yes! Countries with higher life expectancy are also happier.
3. Joining legal rights strength table¶
And the table with a measure of legal rights strength in the country:
df3 = fuzzy_join(
df2,
legal_rights,
left_on="Country",
right_on="Country Name",
match_score=0.45,
)
df3.drop(columns=["Country Name"], inplace=True)
df3.head(3)
Let’s take a look at their correspondance in a figure:
plt.figure(figsize=(4, 3))
fig = sns.regplot(
data=df3,
x="Strength of legal rights index (0=weak to 12=strong)",
y="Happiness score",
lowess=True,
)
fig.set_ylabel("Happiness index")
fig.set_title("Does a country's legal rights strength lead to happiness?")
plt.tight_layout()
plt.show()
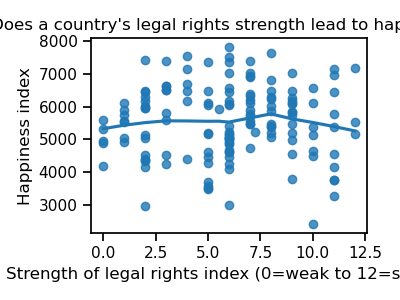
From this plot, it is not clear that this measure of legal strength is linked to happiness.
Great! Our joined table has become bigger and full of useful information. And now we are ready to apply a first machine learning model to it!
Prediction model¶
We now separate our covariates (X), from the target (or exogenous) variables: y
X = df3.drop("Happiness score", axis=1).select_dtypes(exclude=object)
y = df3[["Happiness score"]]
Let us now define the model that will be used to predict the happiness score:
from sklearn import __version__ as sklearn_version
if sklearn_version < "1.0":
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import HistGradientBoostingRegressor
from sklearn.model_selection import KFold
hgdb = HistGradientBoostingRegressor(random_state=0)
cv = KFold(n_splits=2, shuffle=True, random_state=0)
To evaluate our model, we will apply a 4-fold cross-validation. We evaluate our model using the R2 score.
Let’s finally assess the results of our models:
from sklearn.model_selection import cross_validate
cv_results_t = cross_validate(hgdb, X, y, cv=cv, scoring="r2")
cv_r2_t = cv_results_t["test_score"]
print(f"Mean R2 score is {cv_r2_t.mean():.2f} +- {cv_r2_t.std():.2f}")
Out:
Mean R2 score is 0.64 +- 0.02
We have a satisfying first result: an R2 of 0.66!
Data cleaning varies from dataset to dataset: there are as
many ways to clean a table as there are errors. fuzzy_join()
method is generalizable across all datasets.
Data transformation is also often very costly in both time and ressources.
fuzzy_join() is fast and easy-to-use.
Now up to you, try improving our model by adding information into it and beating our result!
Using the FeatureAugmenter() to fuzzy join multiple tables¶
A faster way to merge different tables from the World Bank
to X is to use the FeatureAugmenter().
The FeatureAugmenter() is a transformer that can easily chain joins of tables on
a main table.
Instantiating the transformer¶
y = df["Happiness score"]
We gather the auxilliary tables into a list of (tables, keys) for the tables parameter. An instance of the transformer with the necessary information is:
from dirty_cat import FeatureAugmenter
fa = FeatureAugmenter(
tables=[
(gdppc, "Country Name"),
(life_exp, "Country Name"),
(legal_rights, "Country Name"),
],
main_key="Country",
)
Fitting and transforming into the final table¶
To get our final joined table we will fit and transform the main table (df)
with our create instance of the FeatureAugmenter():
df_final = fa.fit_transform(df)
df_final.head(10)
And that’s it! As previously, we now have a big table ready for machine learning. Let’s create our machine learning pipeline:
from sklearn.pipeline import make_pipeline
from sklearn.compose import make_column_transformer
# We include only the columns that will be pertinent for our regression:
encoder = make_column_transformer(
(
"passthrough",
[
"GDP per capita (current US$)",
"Life expectancy at birth, total (years)",
"Strength of legal rights index (0=weak to 12=strong)",
],
),
remainder="drop",
)
pipeline = make_pipeline(fa, encoder, HistGradientBoostingRegressor())
And the best part is that we are now able to evaluate the parameters of the fuzzy_join().
For instance, the match_score was manually picked and can now be
introduced into a grid search:
from sklearn.model_selection import GridSearchCV
# We will test four possible values of match_score:
params = {"featureaugmenter__match_score": [0.2, 0.3, 0.4, 0.5]}
grid = GridSearchCV(pipeline, param_grid=params)
grid.fit(df, y)
print(grid.best_params_)
Out:
{'featureaugmenter__match_score': 0.2}
The grid searching gave us the best value of 0.5 for the parameter
match_score. Let’s use this value in our regression:
print(f"Mean R2 score with pipeline is {grid.score(df, y):.2f}")
Out:
Mean R2 score with pipeline is 0.83
Great, by evaluating the correct match_score we improved our
results significantly!
Total running time of the script: ( 0 minutes 16.265 seconds)